# this ipython magic command imports pylab and allows the plots to reside within the notebook
%pylab inline
Populating the interactive namespace from numpy and matplotlib
The following examples are based on Florian Lhuillier’s lecture on Matlab which can be found online:
http://geophysik.uni-muenchen.de/~lhuillier/teaching/AD2010/
Pylab continued …¶
Examples based on “… des données sous MATLAB”¶
set_printoptions(precision=2, suppress=True)
Excercise 1¶
aa = loadtxt('dipole_ref.txt')
aa.shape
(21481, 3)
subplot(211)
plot(aa[:,0], aa[:,1])
xlabel("test1")
subplot(212)
plot(aa[:,0], aa[:,2])
xlabel('TEST2')
subplots_adjust(hspace=.5)
Excercise 2¶
aa = loadtxt('dipole_ref.txt')
bb = loadtxt('dipole_pert.txt')
t = bb[:,0];
d = bb[:,2] - interp(t, aa[:,0],aa[:,2]);
semilogy(t, abs(d))
[<matplotlib.lines.Line2D at 0x7f911be2add8>]
Excercise 3¶
d = genfromtxt('spectre.txt', skip_header=1, dtype=None, names=('name', 'number', 'amplitude'))
d
array([(b'a', 1, 9.5960921), (b'b', 2, 9.4450301), (b'c', 3, 7.5000147),
(b'd', 4, 6.7446755), (b'e', 5, 4.7442346), (b'f', 6, 4.3416987),
(b'g', 7, 2.9904522), (b'h', 8, 2.3769902), (b'i', 9, 1.8908512),
(b'j', 10, 1.3387981), (b'k', 11, 1.0044059),
(b'l', 12, 0.79748122), (b'm', 13, 0.5872564),
(b'n', 14, 0.48502449), (b'p', 15, 0.34799267),
(b'q', 16, 0.28167023), (b'r', 17, 0.20974501),
(b's', 18, 0.16570928), (b't', 19, 0.12762576),
(b'u', 20, 0.098378887)],
dtype=[('name', 'S1'), ('number', '<i8'), ('amplitude', '<f8')])
Y=log(d['amplitude']); Y
array([ 2.26, 2.25, 2.01, 1.91, 1.56, 1.47, 1.1 , 0.87, 0.64,
0.29, 0. , -0.23, -0.53, -0.72, -1.06, -1.27, -1.56, -1.8 ,
-2.06, -2.32])
X = array((d['number'], 0*d['number']+1)).T; X
array([[ 1, 1],
[ 2, 1],
[ 3, 1],
[ 4, 1],
[ 5, 1],
[ 6, 1],
[ 7, 1],
[ 8, 1],
[ 9, 1],
[10, 1],
[11, 1],
[12, 1],
[13, 1],
[14, 1],
[15, 1],
[16, 1],
[17, 1],
[18, 1],
[19, 1],
[20, 1]])
X.shape, Y.shape
((20, 2), (20,))
A = lstsq(X, Y)[0]; A
array([-0.25, 2.82])
subplot(211)
plot(d['number'], d['amplitude'])
subplot(212)
semilogy(d['number'], d['amplitude'], 'o')
semilogy(d['number'], exp(X.dot(A)))
[<matplotlib.lines.Line2D at 0x7f9119b37ac8>]
Excercise 4¶
aa = loadtxt('anomalie.txt')
nx = len( unique(aa[:,0])); nx
21
ny = len( unique(aa[:,1])); ny
11
X=aa[:,0].reshape((nx, ny));
Y=aa[:,1].reshape((nx, ny));
data = aa[:,2].reshape((nx, ny));
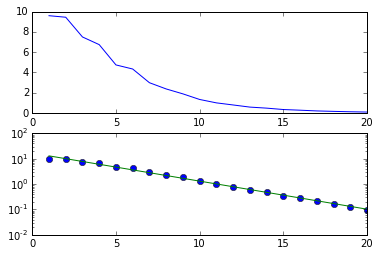
pcolor( X, Y, data)
<matplotlib.collections.PolyCollection at 0x7f9119cbe278>
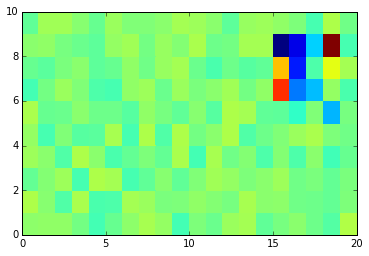
imshow(data.T,
extent=[X.min(), X.max(), Y.min(), Y.max()],
interpolation='gaussian', origin='lower')
colorbar()
<matplotlib.colorbar.Colorbar at 0x7f9119b30ef0>
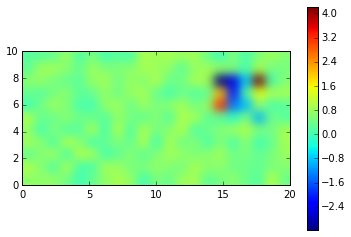
contourf( X, Y, data)
colorbar()
<matplotlib.colorbar.Colorbar at 0x7f9119d93470>
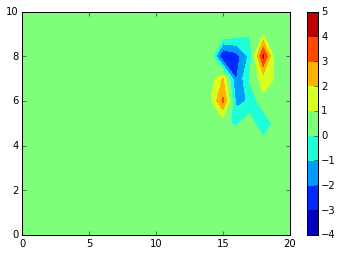
from scipy.ndimage import zoom
data = array(zoom(data,3)).T
contourf(data, extent=[X.min(), X.max(), Y.min(), Y.max()])
colorbar()
#axis("equal")
<matplotlib.colorbar.Colorbar at 0x7f9116fb7be0>
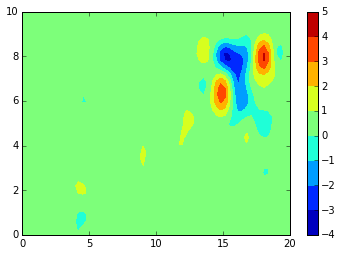
imshow(data,
extent=[X.min(), X.max(), Y.min(), Y.max()],
interpolation='gaussian', origin='lower', aspect=1)
colorbar()
<matplotlib.colorbar.Colorbar at 0x7f9116ef5240>